Quickstart
This section is mainly intended for the installation of G2P and the briefly introduction of several typical usage of G2P to help users get started quickly. Details and other functions of G2P, please see Tutorial
Installation
Installation of G2P container
1. Installation of Singularity (Linux, assuming ubuntu)
1.1 Install system dependencies
$ sudo apt-get update && sudo apt-get install -y \
build-essential \
libssl-dev \
uuid-dev \
libgpgme11-dev \
squashfs-tools \
libseccomp-dev \
wget \
pkg-config \
git \
cryptsetup
1.2 Install GO
$ export VERSION=1.16.4 OS=linux ARCH=amd64 && \ # Replace the values as needed
wget https://dl.google.com/go/go$VERSION.$OS-$ARCH.tar.gz && \ # Downloads the required Go package
sudo tar -C /usr/local -xzvf go$VERSION.$OS-$ARCH.tar.gz && \ # Extracts the archive
rm go$VERSION.$OS-$ARCH.tar.gz # Deletes the ``tar`` file
 Note: You can visit Go Downloads page for suitable installation steps of your operating system.
Note: You can visit Go Downloads page for suitable installation steps of your operating system.
1.3 Install Singularity
You can download SingularityCE from one of the releases. Please visit GitHub release page for the full list. Once you make the decision, you can run the following commands to proceed with the installation (here, we use the version 3.8.1 as example).
$ export VERSION=3.8.1 && # adjust this as necessary \
wget https://github.com/sylabs/singularity/releases/download/v${VERSION}/singularity-ce-${VERSION}.tar.gz && \
tar -xzf singularity-ce-${VERSION}.tar.gz && \
cd singularity-ce-${VERSION}
1.4 Compile singularity
$ ./mconfig && \
make -C builddir && \
sudo make -C builddir install
 Note: If you get an error when compile, please try the following command or refer to Singularity websit for documents.
Note: If you get an error when compile, please try the following command or refer to Singularity websit for documents.
## Motify the file mlocal/frags/go_common_opts.mk in singularity path,find the following line and change it
GOPROXY := https://goproxy.cn,direct
## At the same time, motify the global proxy
go env -w GOPROXY=https://goproxy.cn,direct
1.5 Install Singularity for other operating system Singularity can be installed via Vagrant Boxes for Windows and Intel cores MacOS, the details please refer to Singularity installation guide.
2. Pull G2P container
singularity pull G2P.sif library://mym89757/repo/g2p:latest
Installation of G2P R package
 Note: The following functions, including GSFiltering, GSImputation, GSTransForm, GSRead shown in figure "Overview of G2P" a needs the opearation evironment and stand-alone software on the circle, namely, these function in G2P pcakage can't be performed normally unless the dependencis shown in figure "Overview of G2P" a (upper semicircle) have been correctly installed and configuration. G2P package can be installed for MacOS, Linux and Windows and the functions shown in figure "Overview of G2P" b can run normally.
Note: The following functions, including GSFiltering, GSImputation, GSTransForm, GSRead shown in figure "Overview of G2P" a needs the opearation evironment and stand-alone software on the circle, namely, these function in G2P pcakage can't be performed normally unless the dependencis shown in figure "Overview of G2P" a (upper semicircle) have been correctly installed and configuration. G2P package can be installed for MacOS, Linux and Windows and the functions shown in figure "Overview of G2P" b can run normally.
1. Install G2P through Github
## install dependencies and G2P
install.packages(c("ggplot2","brnn","glmnet","spls","pls","e1071","BGLR","rrBLUP","randomForest","hglm","hglm.data","parallel","pROC","PRROC","STPGA","reshape","reshape2","grid","pbapply","pheatmap","data.table"))
## install package qtlDesign
install.packages(path_of_qtlDesign, repos = NULL, type="source")
## e.g. "/home/Download/qtlDesign_0.941.tar.gz"
require("devtools")
install_github("G2P-env/G2P")
qtlDesign download link
2.Download .tar.gz package and install
## install dependencies and G2P
install.packages(c("ggplot2","brnn","glmnet","spls","pls","e1071","BGLR","rrBLUP","randomForest","hglm","hglm.data","parallel","pROC","PRROC","STPGA","reshape","reshape2","grid","pbapply","pheatmap","data.table"))
## install package qtlDesign
install.packages(path_of_qtlDesign, repos = NULL, type="source")
## e.g. "/home/Download/qtlDesign_0.941.tar.gz"
install.packages("DownloadPath/G2P_1.0.tar.gz")
qtlDesign download link
Usage of G2P container
1. Use G2P for interactive window
$singularity exec G2P.sif R
## library G2P package
library(G2P)
## load example data ##
data("cubic")
## perform genotype to phenotype prediction with all integrated methods ##
C2Pres <- G2P(markers = Markers,data = pheData,trait = "DTT",
modelMethods = c("BayesA", "BayesB", "BayesC", "BL", "BRR",
"RKHS", "RRBLUP","LASSO", "SPLS", "SVC", "SVR", "RFR", "RFC",
"RR", "RKHS", "BRNN"),trainIdx = 1:200,predIdx = 201:400)
2. Use G2P to run Rscript
## perform genotype to phenotype prediction ##
suppressPackageStartupMessages(library(G2P))
print("G2P has been loaded successfully")
## Load example data ##
data("cubic")
## Fit RRBLUP model ##
RRBLUP_model <- GSReModel(markers = Markers[1:200,], pheVal = pheData$DTT[1:200], modelMethods = "RRBLUP")
## Predict 201-400 subset of all example data with already RRBLUP model ##
res <- predictGS(testMat = Markers[201:400,], trainModel = RRBLUP_model, modelMethods = "RRBLUP")
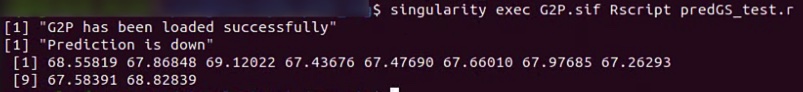
print("Prediction is down")
res[1:10]
Saving above scripts or your own scripts to an R scripts file *.r (e.g. predGS_test.r), Then use G2P container to run it.
$singularity exec G2P.sif Rscript predGS_test.r
3 Use G2P Container to run stand-alone software
- Plink
$singularity exec G2P.sif plink
- Gemma
$singularity exec G2P.sif gemma
- GCTA
$singularity exec G2P.sif GCTA
- TASSEL
$singularity exec G2P.sif Tassel
- VCFtools
$singularity exec G2P.sif vcftools
Usage of G2P pacakge
-
With RStudio

Interface of Rstudio -
From the console
library(G2P)
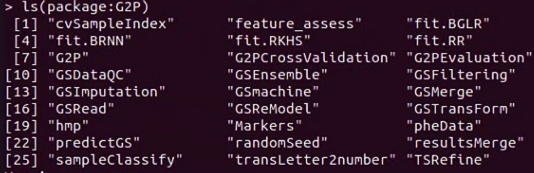
ls("package:G2P")